The method used in this paper is conceptually very similar to a method used in our lab for whole-genome fitness assays. The stated problem is to make single gene knock-out and measure their growth. However, it is not possible to delete the genes one by one in a cell-line. So shRNAs have been the method of choice for this study. They start with an shRNA plasmid library, package the plasmids into a virus and infect the cells to have a library with single genome insertions. In these infected cells, the expression of a gene is knocked down through miRNA like processes.
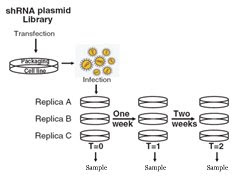 After taking samples at several timepoints of the selection period, they PCR up the constructs, label them and compare them to the unselected library on a microarray chip (see below).
After taking samples at several timepoints of the selection period, they PCR up the constructs, label them and compare them to the unselected library on a microarray chip (see below). Using this approach, the authors identified and later validated essential genes in several human cell-lines.
Using this approach, the authors identified and later validated essential genes in several human cell-lines.

1 comment:
I actually found another paper which uses this method to identify the genes that are not necessary for normal cells but become essential in cancer state (Schlabach et al, 2008. Science 319:620).
Post a Comment